 J Clin Aesthet Dermatol. 2023;16(6):44–45.
J Clin Aesthet Dermatol. 2023;16(6):44–45.
by Jacco Ruiz, BS; Mark Hyde, PhD; Allyson Perry, PhD; and Brook Brouha, MD, PhD
Mr. Ruiz is with the University of California San Diego School of Medicine in San Diego, California. Drs. Hyde and Perry are with DermTech in La Jolla, California. Dr. Brouha is with West Dermatology in San Diego, Californina.
FUNDING: Funding was provided in part by DermTech.
DISCLOSURES: Dr. Brouha is a speaker, PI, and consultant at DermTech, and a speaker and consultant at
Castle Biosciences. Drs. Hyde and Perry are employed by DermTech. Dr. Ruiz reports no conflicts of interest relevant to the content of this article.
ABSTRACT: The clinical evaluation of pigmented lesions represents a ‘high-stakes’ scenario as a missed melanoma can be fatal. Traditional clinical assessment visually sorts pigmented lesions into those that merit a biopsy and those that do not. In our practice there exists a group of lesions judged to not merit biopsy where melanoma, while very unlikely, cannot be excluded with absolute certainty. These ambiguous pigmented lesions (APLs) were often photographed and followed for clinical evolution. This article evaluates the presence of APLs and describes the use of non-invasive genomic testing to sort them. An informal survey using pictures of 10 APLs found that 6 of 8 dermatology providers were unable to identify which were melanomas. Next, our single practice chart review of 1,254 APLs evaluated by non-invasive genomic testing revealed 35 melanomas. All 1,254 were lesions that fell below our biopsy threshold. Non-invasive genomic testing can improve biopsy decisions particularly in clinically indeterminate pigmented lesions. Keywords: Melanoma, nevus, pigmented lesions, genomics, biopsy, PRAME, LINC00518, ambiguous pigmented lesions
During the clinical exam of pigmented lesions, it is common for providers to pause and evaluate some lesions more closely while ultimately choosing not to biopsy them. Because it is not feasible, cost-effective, or beneficial to patients to remove every pigmented lesion, healthcare providers must set a threshold for biopsy. This manuscript evaluates ambiguous pigmented lesions (APLs), pigmented lesions that lie beneath the threshold for biopsy yet cannot be dismissed with absolute certainty (Figure 1). Historically in this practice, these APLs were followed for clinical evolution; now they are tested non-invasively for genomic markers of melanoma. In this report, we review data where we used genomic profiling to test 1,254 APLs on initial presentation instead of following them clinically. Over a 43-month period, we found 35 early-stage melanomas.
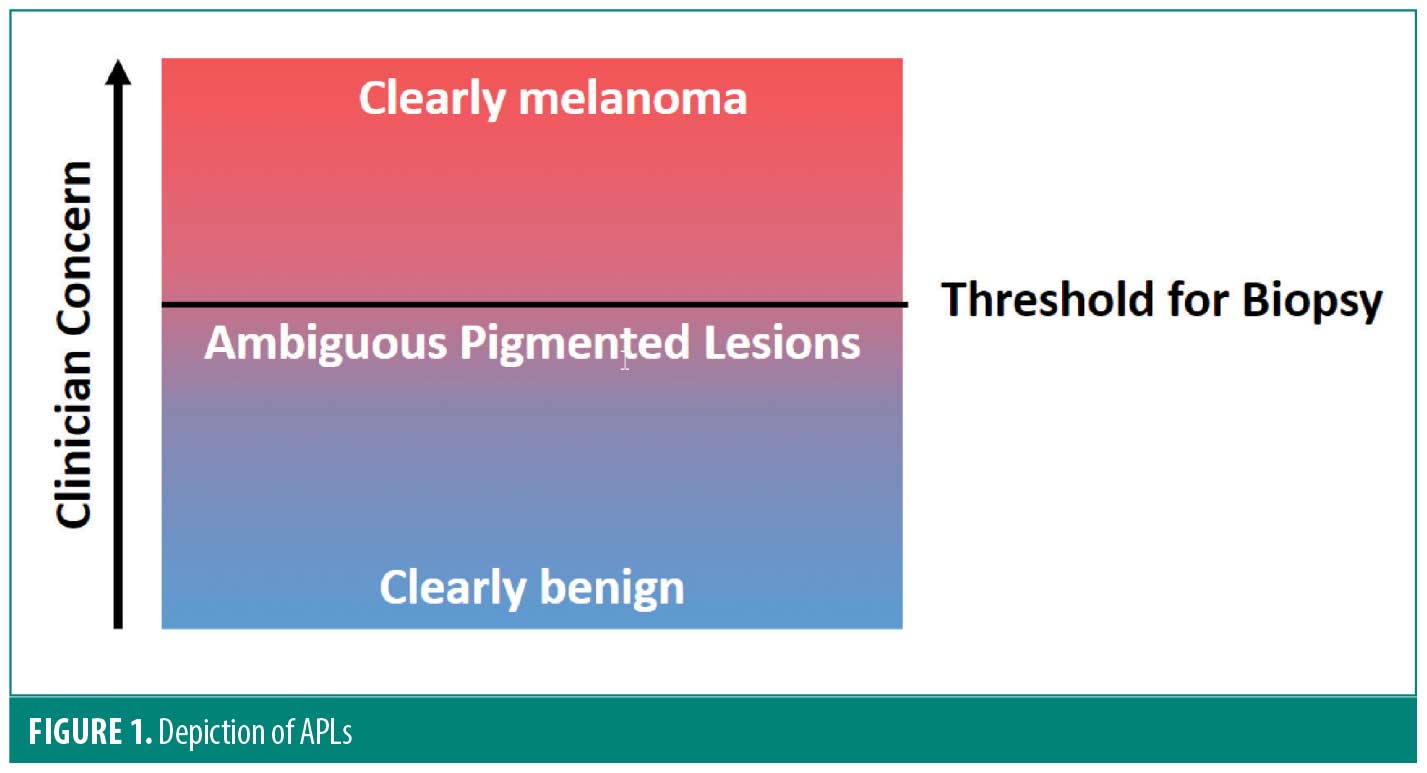
The existence of APLs has been corroborated in discussions with multiple dermatology providers ranging from experienced academic dermatologists to newly trained dermatology physician assistants. Furthermore, a recent study evaluating the utility of dermoscopy to differentiate pigmented lesions showed nearly 60 percent of early melanomas did not exhibit melanoma-specific dermoscopic features, suggesting a gap between malignant transformation and visible changes.1
To demonstrate the difficulty of APLs, a kodachrome-style pilot survey was e-mailed to 13 colleagues. Seven dermatologists and one dermatology physician assistant responded and attempted to classify photographs of 10 APLs as benign or malignant. The 10 lesions were identified from a single practice where five of the lesions were melanoma and five were benign. The top score was 8 of 10 lesions correct with no missed melanomas. One other provider did not miss any melanomas (range 0-3 missed melanomas per provider). The total number of correctly sorted lesions ranged from 3 to 8 per person (average 6.4). Six providers (75%) missed at least one melanoma.
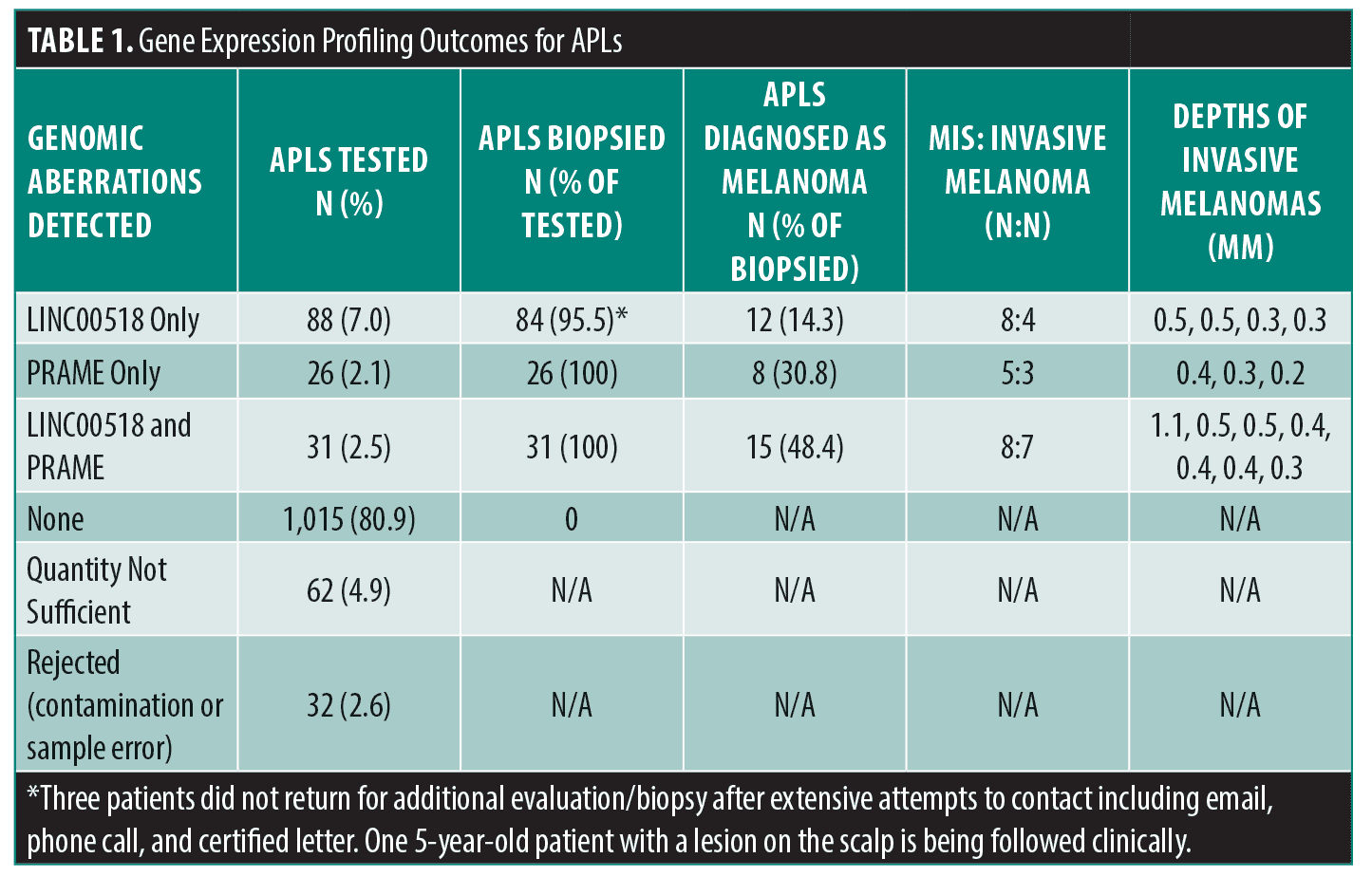
In our practice, APLs are now profiled genomically using non-invasive methods (DermTech, La Jolla, CA) as mentioned in the NCCN melanoma guidelines.2 To define the potential risk of these lesions, we reviewed charts of 1,254 pigmented lesions over 43 months that fell below the biopsy threshold but garnered additional attention. Of 1,254 lesions, 145 (11.6%) expressed genomic markers associated with melanoma, 141 of which were biopsied.3 Thirty-five lesions were found to be melanoma. Twenty-one were melanoma-in-situ and 13 were early-invasive not deeper than 0.5mm. There was one thicker melanoma (1.1mm). One thousand fifteen (80.9%) lesions tested negative, 62 (4.9%) lesions had insufficient yield for genomic evaluation, and 32 (2.6%) lesions were rejected for contamination (hair or blood on the adhesive) or sample error. Of the four not biopsied, three patients did not return for additional evaluation/biopsy after extensive attempts to contact them, including email, phone call, and certified letter. Finally, one 5-year-old patient with a scalp lesion that expressed LINC00518 only has been followed clinically since November 2018 with no clinical changes noted as of January 2022.4 See Table 1 for gene expression patterns and melanoma depths.
The mantra “never miss a melanoma” is rooted in the concept of sensitivity—here, the ability of an office visit to identify individuals with melanoma. Sensitivity of a visit is the number of melanomas found (true positives) divided by the number of melanomas presenting (true positives plus false negatives). To increase the sensitivity of melanoma screenings, we must minimize missed melanomas. The clinical algorithm to visually sort pigmented lesions is a subjective and complex process. Many variables may be considered such as morphology, location, age, and patient preference. APLs may comprise a pool harboring missed melanomas—an opportunity to improve the sensitivity of the clinical exam.
While this is a single practice’s experience, other clinicians might also have difficulty categorizing APLs. The presence and risk of these lesions warrants additional research. While we suspect that pigmented lesion categorization will vary greatly among clinicians, we stratified 1,254 APLs non-invasively and found 35 thin melanomas with only 141 biopsies. In addition, in the 43 months we used genomic profiling to test these lesions, there were no melanomas thicker than 0.7mm in a return patient. The 1,015 negative lesions will continue to be followed clinically.
References
- Babino G, Lallas A, Agozzino M, et al. Melanoma diagnosed on digital dermoscopy monitoring: A side-by-side image comparison is needed to improve early detection. J Am Acad Dermatol. Sep 2021;85(3):619–625.
- National Comprehensive Cancer Network (NCCN). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®). Melanoma: Cutaneous. Version 1.2022. National Comprehensive Cancer Network. Updated December 3, 2021.
- Gerami P, Yao Z, Polsky D, et al. Development and validation of a noninvasive 2-gene molecular assay for cutaneous melanoma. J Am Acad Dermatol. Jan 2017;76(1):114–120 e2.
- Jansen B, Hansen D, Moy R, et al. Gene Expression Analysis Differentiates Melanomas from Spitz Nevi. J Drugs Dermatol. May 01 2018;17(5):574–576.

