 J Clin Aesthet Dermatol. 2021;14(12):44-48.
J Clin Aesthet Dermatol. 2021;14(12):44-48.
by Bijan Safai MD, DSc; Albert G. Wu, MS; and Carl V. Hamby, PhD
Dr. Safai is with the Department of Dermatology, Metropolitan Hospital in New York, New York. Mr. Wu and Dr. Hamby are with New York Medical College School of Medicine in Valhalla, New York.
FUNDING: No funding was provided for this article.
DISCLOSURES: The authors report no conflicts of interest relevant to the content of this article.
ABSTRACT: Background. It is often difficult to accurately predict how a melanoma will progress because melanomas can be so diverse in their genetic and histological makeup.
Objective. We sought to characterize the current state and progression of biomedical markers towards their utilization as prognostic indicators for patients with melanoma.
Methods. A literature search of the research repository databases PubMed and GoogleScholar was conducted using the following inclusion criteria: (1) published within the last 10 years, and (2) use of overall survival, disease progression, or clinical outcome as primary endpoints. Search terms included various permutations of “biomarkers,” “prognostic,” “immunologic,” “serologic,” “visual,” and “melanoma.” Results were evaluated for statistical power, results significance, and experimental design integrity.
Results. The prognostic capabilities of clinical tests for malignant melanoma have made great strides in the last few years, with several serologic and immunohistochemical biomarkers being preliminarily linked to various measures of clinical prognosis. While clinical feasibility of a single sensitive and specific biomarker remains unfeasible, use of select combinations of tested biomarkers remain viable.
Conclusion. Diagnostic and prognostic genetic assays have begun to cross over from research to commercial application, giving physicians additional tools during the early stages of diagnosis to optimize and individualize treatments.
Key words: Immunologic marker, serum marker, screening assay, checkpoint inhibitor, melanoma
Because melanomas can be so diverse in their genetic and histological makeup, it is often difficult to accurately predict how a melanoma will progress. Yet, accurate assessment of this process, including estimating metastatic risk at early tumor stages, is an important factor in reducing patient mortality. Malignant melanomas remain the deadliest form of skin cancer, responsible for an estimated 9,320 deaths in 2018.1 While factors such as age, ethnicity, and tumor location2 can discern broad trends on a macro level, they are not sensitive enough for individual cases. Current standards used as prognostic factors include mitotic rate, Breslow death, and sentinel lymph node (SLN) assessment.3 The American Joint Commission on Cancer (AJCC) includes these variables to stage and classify tumors and generate survival curves for each stage, allowing for more accurate clinical decision-making. However, even in the most recent eighth edition of the AJCC staging system, the lack of novel prognostic biomarkers is strikingly apparent.3,4
In the era of genomics, the use of large-scale screening assays to screen for a predetermined set of markers has largely rendered the idea of a single, tell-all biomarker an archaic notion. Genetic tests are already commercially available to aid clinicians with melanoma staging and diagnosis. Treatment options for melanoma have also evolved, with checkpoint inhibitors improving patient survival rates5 for late-stage melanoma. However, therapeutic benefits vary between individuals, as studies have expressed difficulty in anticipating patient responses6 as well as the insufficiency of conventional response criteria for predicting therapeutic benefit.7 This variation has driven the search for biomarkers which can serve as indicators of patient response to therapy. Recently, a number of studies have been able to uncover several genetic and protein markers that show statistically significant associations with mortality, improved clinical outcome, and melanoma progression. We aim to summarize and highlight these markers, which have the potential to aid in the development of individualized treatments for malignant melanomas in the future.
Methods
A literature search of the research repository databases PubMed and GoogleScholar was conducted using the following inclusion criteria: (1) published within the last 10 years,and (2) use of overall survival, disease progression, or clinical outcome as primary endpoints. Search terms included various permutations of “biomarkers,” “prognostic,” “immunologic,” “serologic,” “visual,” and “melanoma.” Results were evaluated for statistical power, results significance, and experimental design integrity.
Results
Immunohistochemical markers. The wave of retrospective reviews in the last five years has brought up many immunohistochemical biomarkers associated with the prognosis of malignant melanomas, many of which are summarized in Table 1. It is important to stress that many of these studies were limited by having small cohorts, using single centers, or studying rare variants of malignant melanoma. These initial findings will need to be replicated and expanded for any definitive claims to be made; however, they add to the pool of potential markers and therapeutic targets to be tested for future use.
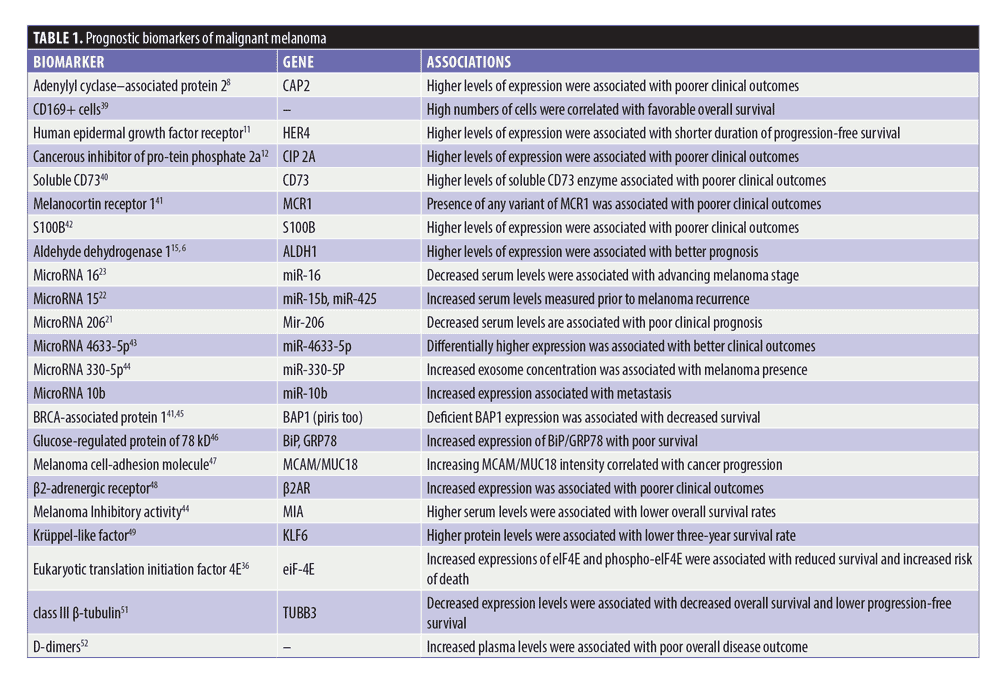
Adenylate cyclase–associated protein (CAP2). CAP2 is an actin monomer binding protein that plays a regulatory role in actin assembly and disassembly in the cell.8 It has been shown to play roles in the proliferation and migration of breast cancer cells and is overexpressed in hepatocellular carcinoma.9 More recently, a retrospective study of 50 patients with metastatic melanoma found that CAP2 overexpression was significantly associated with increased tumor thickness and poorer clinical outcomes. In addition, gene expression increased in a stepwise pattern as melanoma stage advanced, which opens the possibility of it being a marker of cancer progression.8
Epidermal growth factor receptor (EGFR). The EGFR family, consisting of EGFR/HER1, HER2, HER3, and HER4, regulates several cellular processes important for growth and survival, including cell division, apoptosis, and migration.10 HER4 expression is normally associated with antiproliferative and pro-apoptotic activity. It is also frequently mutated in malignant melanomas (19%), raising the question of whether it could be used to inform patient prognosis or identify malignancies earlier in the clinical setting. In two separate single-center retrospective studies, HER4 expression levels were found to be significantly associated with patient survival and prognosis.10,11 While HER4 can have both oncogenic and tumor-suppressor properties, both studies found HER4 expression to be oncogenic in malignant melanoma, with high expression levels significantly associated with shorter overall progression-free survival.10,11 Patients positive for HER4 expression were also shown to have lower overall survival than those who were HER4-negative.10,11 These studies point to HER4 as a potential predictor of patient prognosis or at least a molecular therapeutic target.
Cancerous inhibitor of protein phosphate 2a (CIP2A). CIP2A normally regulates C-myc and AKT through its inhibition of protein phosphatase 2A, but it is also an oncogene that promotes tumor transformation and cancer progression.12 CIP2A overexpression has been noted in chronic myelogenous leukemia, ovarian cancer, prostate cancer, and colon cancers, among others.12,13 It has also been recommended as a therapeutic target for head and neck squamous cell carcinoma and triple negative breast cancer.13 Recently, CIP2A was also shown to be overexpressed in malignant melanomas, with high expression significantly associated with poor patient survival and Breslow thickness. Interestingly, there is a distinction between nuclear overexpression of CIP2A, which is associated with poorer overall survival, and cytoplasmic overexpression of CIP2A, which was reported to be correlated with longer relapse-free survival.12 While further research needs to be conducted, these results at least corroborate the idea that CIP2A levels could be used as a predictor of overall survival in melanomas.
Aldehyde dehydrogenases (ALDHs). ALDHs play an important role in tumor cells, neutralizing aldehydes by converting them to carboxylic acids to prevent toxic buildup.14 Previous research has shown that an overexpression of ALDH variants is associated with tumorigenic and chemotherapy-resistant melanoma cells and that ALDH activity is epigenetically upregulated in melanoma cells.15 In addition, it has been reported that silencing of the ALDH1A isozyme leads to cell cycle arrest, reduced tumorigenesis, and drug-sensitized melanoma cells.15 It is interesting then that high levels of ALDH1 activity are associated with better patient outcomes, including decreased rates of melanoma-specific death.16 This is not the only cancer with this inverse relationship, and the variability might be caused by the many different roles ALDH1 could play in each tumor.
Serum markers. Serologic markers are extremely appealing as biomarker candidates as testing is less invasive and has relatively fast turnaround times.17,18 The identification of new metabolites, antigens, and enzymes that could be used as a marker of disease progression and predictors of patient outcomes has driven research over the last few years. Another focus of research has been the re-evaluation of classic biomarkers for their clinical utility and prognostic strength.
RNAs. MicroRNAs (miRNAs) are responsible for attenuating gene expression post-translationally and play many roles in cellular processes, including immune response, apoptosis, and proliferation. They have also been implicated in tumorigenesis and metastasis.19,20 Recently, several serum miRNAs have been reported to be associated with cancer progression and patient prognosis, including miRNA 20621, miR-15022, miR-42522, and miR-1623. When used in an assay, it was reported that a certain miRNA signature was predictive of melanoma recurrence—a finding that could aid in the clinical management of patients at the time of diagnosis.22 Factors implicated in mRNA regulation, such as translation initiation factor 4E (eIF4E), have also been associated with reduced survival.49 Because of the stability of blood and the standardization of assays, blood-based miRNA assays may be low-cost, effective prognostic predictors for malignant melanomas in the future.
5-S-cysteinyl-dopa (5-S-CD) and lactate dehydrogenase (LDH). RNA and genetic markers are not the only category of markers being evaluated for their prognostic capability. 5-S-CD and LDH are a metabolite and enzyme routinely used as serum biomarkers in Japan.24 In one large-scale retrospective review, it was suggested that 5-S-CD levels could significantly distinguish between patients with good and poor prognoses and that elevated levels reflect disease progression.24 LDH has been suggested as a prognostic indicator for some time, but results have been controversial. A recent meta-analysis attempted to further clarify the relationship between LDH and melanoma and found that the high LDH levels could be a predictor of poor prognosis among melanoma patients.25
Biomarker applications in malignant melanoma treatments. Recently, the number of treatments for malignant melanoma has expanded. Immune checkpoint inhibitors, which harness T-cells to amplify immune system responses against tumors, have been extensively researched due to promising long-term clinical results. However, obstacles such as developed resistance and limited treatment scope make traditional chemotherapy the main option for many patients.
Programmed death ligand 1 (PD-L1). PD-L1 is a ligand which binds to programmed death 1 (PD-1) immunoglobin to suppress T-cell activity.26–29 The ligand is expressed in both hematopoietic and nonhematopoietic tissues as well as in tumor cells. It is thought to help tumor cells evade T-cell–stimulated immunological responses; in fact, PD-L1 expression in desmoplastic melanoma is associated with tumor progression.27 Anti–PD-1 inhibitors, such as nivolumab and pembrolizumab, have been shown to have antitumoral responses for many cancers, but the need for indicators of progression remains a focus of research.29 It was recently reported that elevated serum ecto-5’-nucleotidase (CD73) levels were significantly associated with poorer clinical outcomes among patients receiving the PD-L1 inhibitor nivolumab.28,29 PD-L1 itself has also been tested as a marker for patient progression during immune checkpoint inhibitor therapy, and there have been reported associations between higher starting levels of PD-L1 and worse clinical outcomes.26 PDL expression levels have also been used to compare the effectiveness of therapies, which could be valuable in the future as a patient-screening method to individualize treatment options.
Melanocortin 1 receptor (MC1R) and Beta2-adrenergic receptor (Beta2AR). The presence of MC1R,47 a G protein–coupled receptor that plays a role in skin pigmentation, was associated with poorer outcomes in patients. The serological biomarker S100B has been one of the most analyzed prognostic indicators, but its clinical application is limited due to its false-positive rate. Contrastingly, Beta2AR48 is another G protein–coupled receptor found to be an independent prognostic factor predicting poorer survival and was associated with tumor thickness, ulceration, disease stage, and cell proliferation.
Other serological biomarkers. The tumor-suppressor genes BRAFV600E and BRCA-associated protein 1 (BAP1) have also been found in combination to be a consistent immunohistologic signature of a specific type of BAP1 melanocytic lesions. The glucose-regulated protein of 78kd (GRP 78) was also found by Ishikawa et al46 to be an independent prognostic factor for poor survival; however, its elevation is not specific to malignant melanoma. The melanoma cell adhesion molecule was known to be associated with relapse in melanoma. In a modest 175-patient study, Rapanotti et al47 found it was a marker of progression even in early stages of melanoma.
Chemotherapies. Oxaliplatin chemotherapy is an option for colorectal cancer that cross-links in DNA to inhibit replication and transcription, and advances are still being made in what it can treat. It was recently reported that XPF protein levels could be used as a patient screening option for melanoma sensitivity to oxaliplatin therapy.30 Even as novel malignant melanoma treatments are developed, biomarkers still play a role in making traditional therapies more specific to the patients they are most effective for. Another melanoma drug currently in clinical trials, sunitinib, recently had a biomarker analysis performed. Although a couple of candidates (VEGFR1 and P1GF) had correlations with overall survival, no significant associations were made in the study.31 Predictive biomarkers for the use of patient selection for maximal therapeutic benefit from a treatment are still an extremely important topic of research for the future.
Biomarkers and screening assays. Biomarker assays and assessments that integrate multiple prognostic factors are an aspect of research focusing on combining and weighting current data to make the most accurate predictions concerning malignant melanoma diagnoses and prognoses.2,18 Because melanomas often have different combinations of mutations and different levels of expression changes, these assays may be able to somewhat make up for skews in small patient cohorts and prove more consistent than tests based on a single biomarker.
Large-scale genomic sequencing assays are also technologically feasible and can be used on large tissue collections and databases to detect biomarker candidates and genetic variants of melanomas, adding to the available information for clinical use. The most available example of this is Decisiondx (Castle Biosciences; Friendswood, Texas), a commercially available 31-gene expression profiling test, which uses 28 discriminating genes and three control genes to analyze a fixed tissue sample of the tumor.32 Multiple studies33,34 have supported its prognostic utility in aiding in early-stage identification of patients with a higher risk of relapse.
Other biomarker panels with novel genetic candidates are also in the process of being developed. A recent study by Vendittelli et al35 analyzed several biomarker candidates from patient serum samples using quantitative polymerase chain reaction. The analysis was able to compare many biomarkers simultaneously and single out one which had expression levels correlated with Breslow thickness and diagnostic capacity.35,36 Another study used a next-generation sequencing (NGS) panel to sequence several melanoma samples, among other cancers, for somatic variations. Researchers were able to find common expression patterns among the variants, with NGS providing high sequencing fidelity and coverage.37 Panels screening multiple melanoma microRNAs have been conducted as well.38 Other research has pointed to using combined bisulfite restriction analysis assays to identify epigenetic markers in malignant melanoma.39 Overall, improvements in analysis and assay technology have allowed for entire panels of tissue to be analyzed for markers, giving researchers a powerful way by which to study large patient cohorts in the future.
Conclusion
The prognostic capabilities of clinical tests for malignant melanoma have made great strides over the last few years. Diagnostic and prognostic genetic assays have begun to cross over from research to commercial application, giving physicians additional tools during the early stages of diagnosis to optimize and individualize treatments. A significant number of biomarkers from many sources remain to be analyzed and tested, with the potential to improve upon and further optimize current tests. As novel treatments for melanoma continue to be developed, innovation must continue to isolate biomarkers which can help track their efficacy.
References
- Melanoma – Statistics. (2012, June 25). Retrieved from https://www.cancer.net/cancer-types/melanoma/statistics
- Rozeman EA, Dekker TJA, Haanen JBAG, Blank CU. Advanced Melanoma: Current Treatment Options, Biomarkers, and Future Perspectives. American Journal of Clinical Dermatology. 2018;19(3), 303–317.
- Keung EZ, Gershenwald JE. The eighth edition American Joint Committee on Cancer (AJCC) melanoma staging system: implications for melanoma treatment and care. Expert Review of Anticancer Therapy. 2018;18(8):775–784.
- Grob JJ, Schadendorf D, Lorigan P. Eighth American Joint Committee on Cancer (AJCC) melanoma classification: Let us reconsider stage III. European Journal of Cancer. 2018;91: 168–170.
- Hogan SA, Levesque MP, Cheng PF. Melanoma Immunotherapy: Next-Generation Biomarkers. Frontiers in Oncology. 2018;8:178.
- Pennock GK, Waterfield W, Wolchok JD. Patient Responses to Ipilimumab, a Novel Immunopotentiator for Metastatic Melanoma: How Different are these From Conventional Treatment Responses? American Journal of Clinical Oncology. 2012;35(6):606–611.
- Hodi FS, Hwu WJ, Kefford R, et al. Evaluation of Immune-Related Response Criteria and RECIST v1.1 in Patients With Advanced Melanoma Treated With Pembrolizumab. Journal of Clinical Oncology. 2016;34(13):1510–1517.
- Masugi Y, Tanese K, Emoto K, et al. Overexpression of adenylate cyclase-associated protein 2 is a novel prognostic marker in malignant melanoma: CAP2 overexpression in melanoma. Pathology International. 2015;65(12):627–634.
- Yu XF, Ni QC, Chen JP, et al. Knocking down the expression of adenylate cyclase-associated protein 1 inhibits the proliferation and migration of breast cancer cells. Experimental and Molecular Pathology. 2014;96(2):188–194.
- Zhu W, Li S, Zou B, et al. Expressions and clinical significance of HER4 and CD44 in sinonasal mucosal malignant melanoma. Melanoma Research. 2018;28(2):105–110.
- Nielsen TO, Poulsen SS, Journe F, et al. HER4 and its cytoplasmic isoforms are associated with progression-free survival of malignant melanoma. Melanoma Research. 2014;24(1):88–91.
- Shi F, Ding Y, Ju S, et al. Expression and prognostic significance of CIP2A in cutaneous malignant melanoma. Biomarkers. 2014;19(1):70–76.
- Cellular localization of CIP2A determines its prognostic impact in superficial spreading and nodular melanoma. Cancer Medicine. 4(6):903–913.
- Kozovska Z, Gabrisova V, Kucerova L. Malignant melanoma: diagnosis, treatment and cancer stem cells. Neoplasma. 2016;63(04):510–517.
- Pérez-Alea M, McGrail K, Sánchez-Redondo S, et al. ALDH1A3 is epigenetically regulated during melanocyte transformation and is a target for melanoma treatment. Oncogene. 2017;36(41), 5695–5708.
- Taylor LA, Abraham RM, Tahirovic E, et al. High ALDH1 expression correlates with better prognosis in tumorigenic malignant melanoma. Modern Pathology. 2017;30(5), 634–639.
- Abbas O, Miller DD, Bhawan J. Cutaneous Malignant Melanoma: Update on Diagnostic and Prognostic Biomarkers. The American Journal of Dermatopathology. 2014;36(5), 363–379.
- Weinstein D, Leininger J, Hamby C, Safai B. Diagnostic and Prognostic Biomarkers in Melanoma. The Journal of Clinical and Aesthetic Dermatology. 2014;7(6):13–24.
- Ross CL, Kaushik S, Valdes-Rodriguez R, Anvekar R. MicroRNAs in cutaneous melanoma: Role as diagnostic and prognostic biomarkers. Journal of Cellular Physiology. 2018;233(7), 5133–5141.
- Saldanha G, Elshaw S, Sachs P, et al. microRNA-10b is a prognostic biomarker for melanoma. Modern Pathology. 2016;29(2):112–121.
- Tian R, Liu T, Qiao L, Gao M, Li J. Decreased serum microRNA-206 level predicts unfavorable prognosis in patients with melanoma. International Journal of Clinical and Experimental Pathology. 2015;8(3):3097–3103.
- Fleming NH, Zhong J, da Silva IP, et al. Serum-based miRNAs in the prediction and detection of recurrence in melanoma patients: Prognostic Serum miRNAs in Melanoma. Cancer. 2015;121(1):51–59.
- Guo S, Guo W, Li D, et al. Serum miR-16: A Potential Biomarker for Predicting Melanoma Prognosis. Journal of Investigative Dermatology. 2016;136(5):985–993.
- Umemura H, Yamasaki O, Kaji T, et al. Usefulness of serum 5-S-cysteinyl-dopa as a biomarker for predicting prognosis and detecting relapse in patients with advanced stage malignant melanoma. The Journal of Dermatology. 2017;44(4):449–454.
- Health Reference Center Academic – Document – Findings from Department of Dermatology Update Knowledge of Melanoma (Serum lactate dehydrogenase is a predictor of poor survival in malignant). (n.d.). Retrieved January 20, 2019, from http://go.galegroup.com.lproxy.nymc.edu
- Zhou, J., Mahoney, K. M., Giobbie-Hurder, A. Soluble PD-L1 as a Biomarker in Malignant Melanoma Treated with Checkpoint Blockade. Cancer Immunology Research. 2017;5(6): 480–492.
- Kraft S, Fernandez-Figueras MT, Richarz NA, et al. PDL1 expression in desmoplastic melanoma is associated with tumor aggressiveness and progression. Journal of the American Academy of Dermatology. 2017;77(3):534–542.
- Lutzky, J. Checkpoint inhibitors in the treatment of cutaneous malignant melanoma. Chinese Clinical Oncology. 2014;3(3):12.
- Krähenbühl L, Goldinger SM, Mangana J, et al. A Longitudinal Analysis of IDO and PDL1 Expression during Immune- or Targeted Therapy in Advanced Melanoma. Neoplasia. 2018;20(2): 218–225.
- Hatch SB, Swift LP, Caporal S, et al. XPF protein levels determine sensitivity of malignant melanoma cells to oxaliplatin chemotherapy: Suitability as a biomarker for patient selection: Sensitivity of MM cells to oxaliplatin chemotherapy. International Journal of Cancer. 2014;134(6):1495–1503.
- Decoster L, Broek IV, Neyns B, et al. Biomarker Analysis in a Phase II Study of Sunitinib in Patients with Advanced Melanoma. Anticancer Res. 2015;35(12):6893–6899.
- DecisionDx-Melanoma: New genomic test for cutaneous melanoma. (n.d.). Retrieved from Skin Melanoma Test website: https://skinmelanoma.com/managed-care-professionals/decisiondx-melanoma-summary/
- Cook RW, Middlebrook B, Wilkinson J, et al. Analytic validity of DecisionDx-Melanoma, a gene expression profile test for determining metastatic risk in melanoma patients. Diagnostic Pathology. 2018;13:13.
- Keller J, Schwartz TL, Lizalek JM, et al. Prospective validation of the prognostic 31-gene expression profiling test in primary cutaneous melanoma. Cancer Medicine. 2019;8(5):2205–2212.
- Vendittelli F, Paolillo C, Autilio C, Lavieri, M, et al. Absolute quantitative PCR for detection of molecular biomarkers in melanoma patients: A preliminary report. Clinica Chimica Acta. 2015;444:242–249.
- Fisher KE, Zhang L, Wang J, et al. Clinical Validation and Implementation of a Targeted Next-Generation Sequencing Assay to Detect Somatic Variants in Non-Small Cell Lung, Melanoma, and Gastrointestinal Malignancies. The Journal of Molecular Diagnostics. 2016;18(2):299–315.
- Stark MS, Klein K, Weide B, et al. The Prognostic and Predictive Value of Melanoma-related MicroRNAs Using Tissue and Serum: A MicroRNA Expression Analysis. EBioMedicine. 2015;2(7):671–680.
- Kaehler KC, Politz O, Henderson D, et al. Novel DNA methylation markers with potential prognostic relevance in advanced malignant melanoma identified using COBRA assays. Melanoma Research. 2015;25(3):225–231.
- Saito Y, Ohnishi K, Miyashita A, et al. Prognostic Significance of CD169+ Lymph Node Sinus Macrophages in Patients with Malignant Melanoma. Cancer Immunology Research. 2015;3(12):1356–1363.
- Morello S, Capone M, Sorrentino C, et al. Soluble CD73 as biomarker in patients with metastatic melanoma patients treated with nivolumab. Journal of Translational Medicine. 2017;15(1).
- Guida M, Strippoli S, Ferretta A, et al. Detrimental effects of melanocortin-1 receptor (MC1R) variants on the clinical outcomes of BRAF V600 metastatic melanoma patients treated with BRAF inhibitors. Pigment Cell & Melanoma Research. 2016;29(6):679–687.
- Gebhardt C, Lichtenberger R, Utikal J. Biomarker value and pitfalls of serum S100B in the follow-up of high-risk melanoma patients: Value and pitfalls of serum S100B in melanoma. JDDG: Journal Der Deutschen Dermatologischen Gesellschaft. 2016;14(2):158–164.
- Zou B, Zhu W, Liu H, et al. Identification and Functional Evaluation of miR-4633-5p as a Biomarker and Tumor Suppressor in Metastatic Melanoma. Cellular Physiology and Biochemistry. 2018;49(4):1364–1379.
- Su B, Zhou S, Gan C, Zhang, X. MiR-330-5p regulates tyrosinase and PDIA3 expression and suppresses cell proliferation and invasion in cutaneous malignant melanoma. Journal of Surgical Research. 2016;203(2):434–440.
- Piris A, Mihm MC, Hoang MP. BAP1 and BRAFV600E expression in benign and malignant melanocytic proliferations. Human Pathology. 2015;46(2):239–245.
- Shimizu A, Kaira K, Yasuda M, et al. Clinical and Pathological Significance of ER Stress Marker (BiP/GRP78 and PERK) Expression in Malignant Melanoma. Pathology & Oncology Research, 2017;23(1):111–116.
- Rapanotti MC, Suarez Viguria TM, Costanza G, et al. Sequential molecular analysis of circulating MCAM/MUC18 expression: a promising disease biomarker related to clinical outcome in melanoma. Archives of Dermatological Research. 2014;306(6):527–537.
- Shimizu A, Kaira K, Mori K, et al. Prognostic significance of ?2-adrenergic receptor expression in malignant melanoma. Tumor Biology. 2016;37(5):5971–5978.
- Cai D, Zhao J, Sun Q. Krüppel-like factor 6 in the progression and prognosis of malignant melanoma. Journal of International Medical Research. 2014;42(1):184–190.
- Carter JH, Deddens JA, Spaulding IV, et al. Phosphorylation of eIF4E serine 209 is associated with tumour progression and reduced survival in malignant melanoma. British Journal of Cancer. 2016;114(4):444–453.
- Shimizu A, Kaira K, Yasuda, M, et al. Decreased expression of class III ?-tubulin is associated with unfavourable prognosis in patients with malignant melanoma. Melanoma Research. 2016;26(1):29–34.
- Desch A, Gebhardt C, Utikal J, Schneider, SW. D-dimers in malignant melanoma: Association with prognosis and dynamic variation in disease progress: D-dimers in malignant melanoma. International Journal of Cancer. 2017;140(4):914–921.

